Discussion
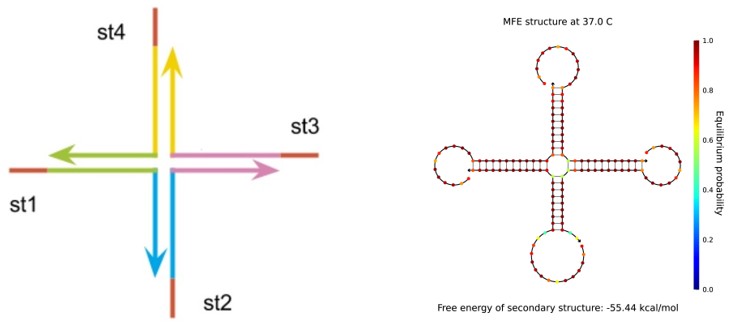
The main point of our experiment is as follows. We synthesised the gel with st1, st2, st3 and st4 (Fig.1)
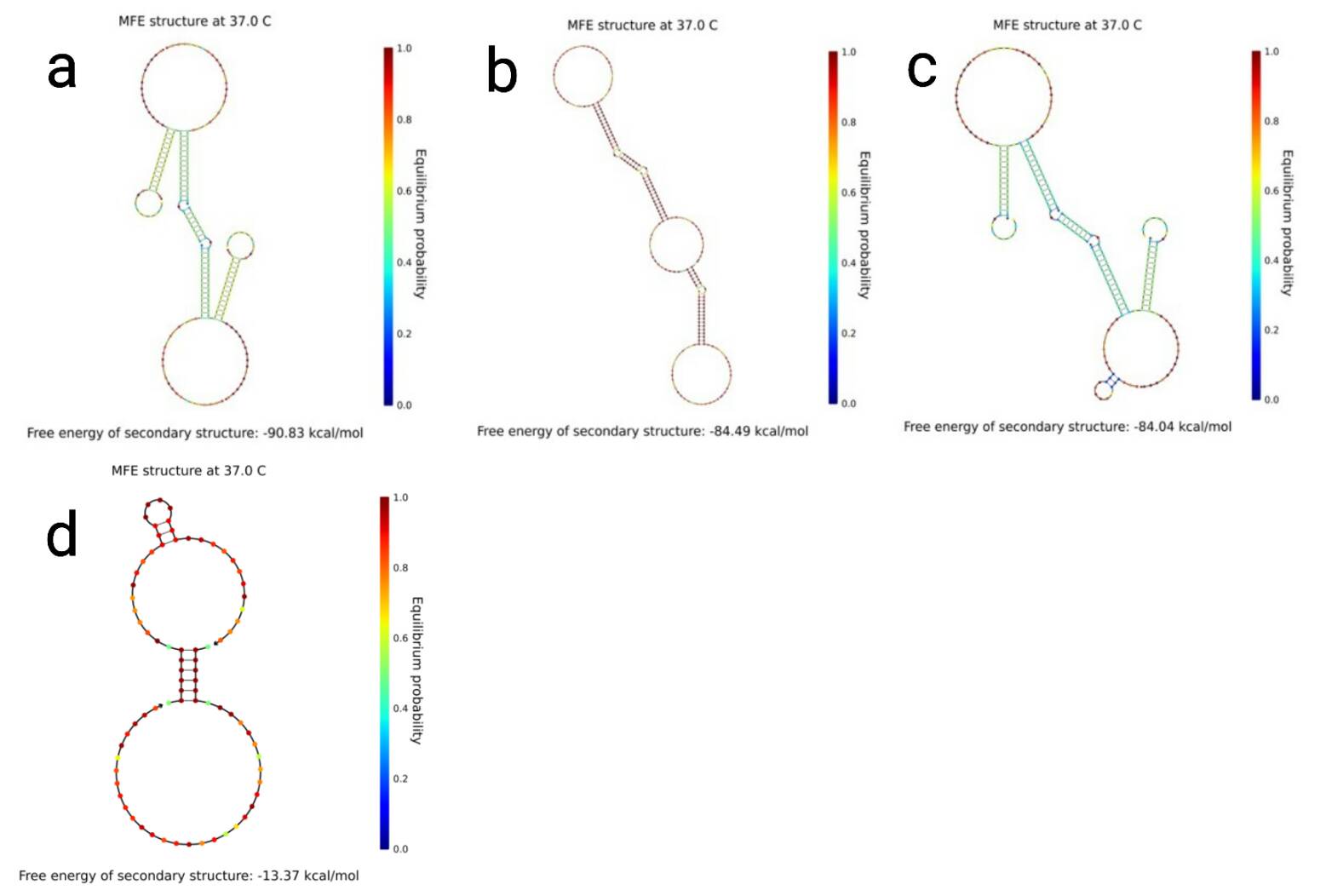
We checked the formation of the gel by annealing strands st1, st2, st3 and st4 by Experiment 1. Based on the results we carried out Experiments 4-1, 2 and confirmed that the gel does not form only when both (st2 t8)* and (st4 t8)* exist in a sufficient concentration. We confirmed the formation of the gel by dropping small amounts of ink on the sample, and then looking at whether the ink diffused or stayed on top of the sample. Although we can check the formation of the gel by this method, we cannot measure the density of the gel quantitatively. If we could think of a way to measure the density quantitatively, we can see the relationship between the concentration of each strands and the density of the gel in Experiment 2, the combinations of strands and the density of the gel in Experiment 3, the impact on the density of the gel when only either (st2 t8)* or (st4 t8)* is added in Experiment 4-2. Results of Experiment 3 and Experiment 4-2 show that only combinations: (Fig2-a) st1, st2, st3; (Fig2-b) st1, st2; (Fig2-c) st1, st3, st2 t8; and (Fig2-d) st1, st3, st4 t8 form a gel. Furthermore combination st1, st3 did not form a gel. From these results, we can infer that when strands exist only diagonally it do not form a gel. We used NUPACK(http://www.nupack.org/) to see the structure which is formed in the combinations mentioned above. (Fig.2)
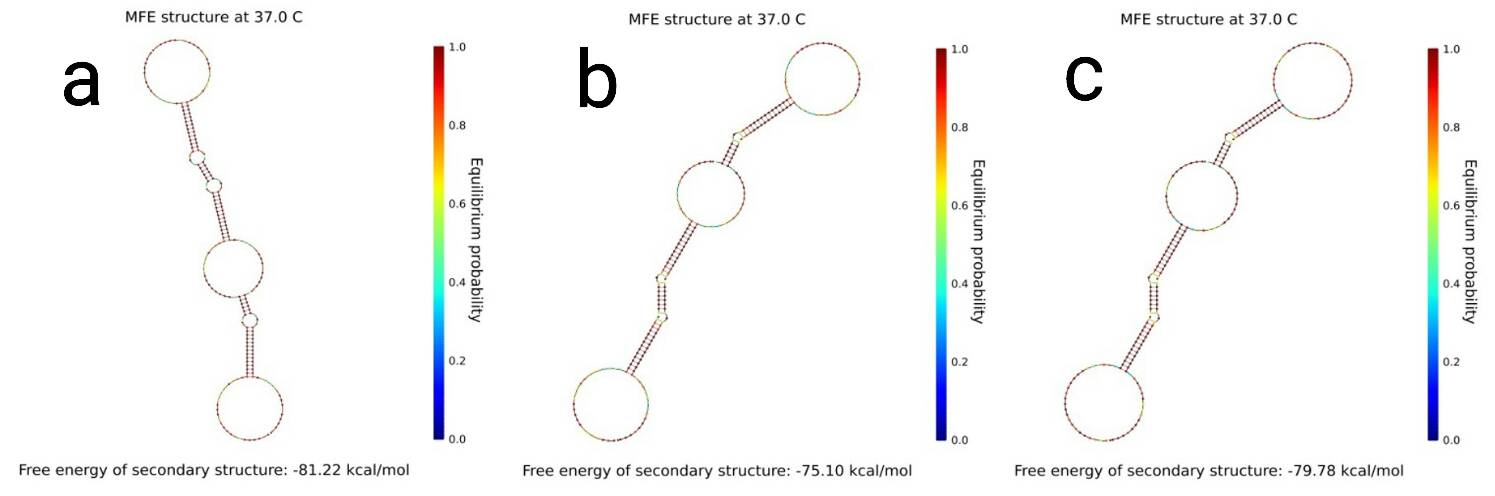
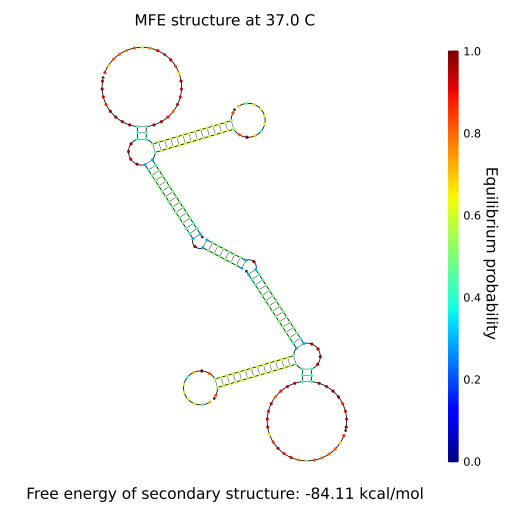
As we can see, the combinations that form a gel bonds in a long line, while the combination that does not form a gel is scattered in a single or bimolecular cluster. Using these, we predicted if gels form in other combinations we did not experiment. (Fig.3, 4 and 5) Our gel has a function of an AND gate in DNA computing. Enzyme-free logic circuits[1] and Reversible logic circuits[2] are famous for AND gates. The former releases a single stranded DNA, while the latter exposes a single stranded DNA sequence as output, but we use the gel-sol transition as output which is new from previous research. [1] Seelig, G., Soloveichik, D., Zhang, D.Y., and Winfree, E., Enzyme-Free Nucleic Acid Logic Circuits, Science 314(5805):1585–1588, 2006. [2] A. J. Genot, J. Bath , and A. J. Turberfield, Reversible Logic Circuits Made of DNA. J. Am. Chem. Soc., 133(50):20080–20083, 2011.